When it comes to ALS research, small breakthroughs can open big doors. That’s exactly what’s happening through a groundbreaking project led by Dr. Brent Harris (Georgetown University) and Dr. Robert Bowser (Barrow Neurological Institute), supported by Target ALS and the Chan Zuckerberg Initiative.
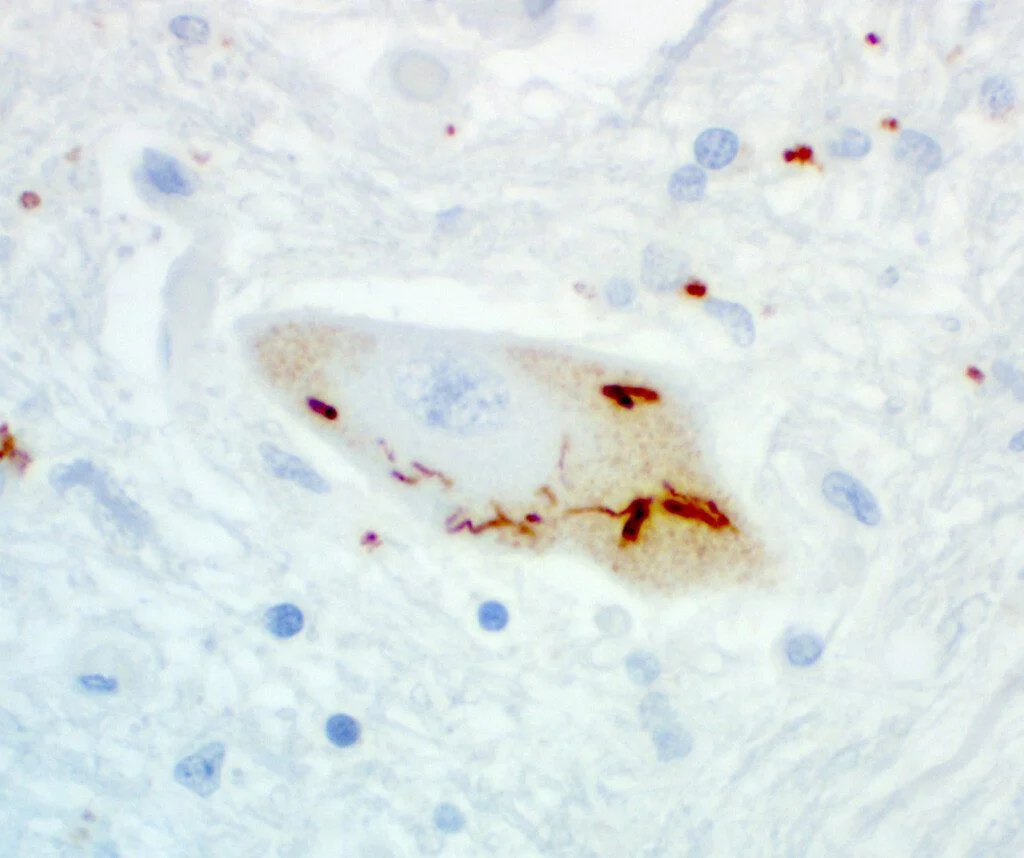
Dr. Harris and Dr. Bowser, heads of Target ALS’s large Post-mortem tissue Core, have dedicated years to stewarding this 500-case repository of human postmortem tissue for the advancement of ALS research. Now, for the first time, hundreds of postmortem ALS and FTD tissue samples have been uniformly stained for phosphorylated TDP-43, a protein known to clump in brain and spinal cord cells and a central hallmark of ALS pathology, using an antibody developed by Dr. Len Petrucelli’s lab. Dr. Harris and Dr. Bowser’s teams are staining and digitizing images of brain and spinal cord sections, pairing them with extensive genetic and clinical metadata, and making them freely accessible through the Target ALS Data Engine.

“No one’s really done this in ALS before,” Dr. Bowser explained. “By moving beyond subjective scoring, we’re creating a quantitative, unbiased view of TDP-43 pathology across cases.”
Why This Matters
Traditionally, neuropathologists scored stained slides on a rough four-point scale: none, mild, moderate, or severe. That subjectivity meant “a lot” of TDP-43 could look different depending on who was judging. With whole-slide imaging and digitization, researchers can view the staining directly, selecting cases with the specific pathology patterns that matter most for their experiments.
And because each image is tied to rich metadata, including long-read sequencing and clinical history, the potential for discovery multiplies. “It’s a holistic approach,” said Dr. Harris. “We’re connecting what we see under the microscope with what we know at the genome, transcriptome, and protein levels and linking it back to patient disease.”

Enter Machine Learning
The next frontier is applying machine learning. In collaboration with Dr. Finkbeiner’s UCSF lab algorithms are being trained to quantifyTDP-43 pathology across thousands of slides. Automated quantification is not only more accurate and reliable but much faster, reducing weeks to months of time-intensive work it would take to try to score each section by hand to a few days.
As Dr. Steve Finkbeiner explained, traditional neuropathology often zeroes in on the “worst” regions and provides semi-quantitative scores, but AI/ML can analyze all tissue across multiple regions, something human pathologists simply don’t have the time to do. And by training models on consensus input from multiple experts, this approach also helps overcome the variability that exists between individual pathologists, increasing reproducibility.
Not only that but AI can detect new patterns of TDP43 pathology and new relationships between TDP43 and other aspects of pathology, to revolutionize the way histopathology is used in research. The proof-of-concept is simple but powerful: once validated, this approach can scale across the entire Target ALS collection.
As Dr. Finkbeiner noted, “I can’t think of another example of a tissue collection that is publicly available and where AI/ML is being applied. This could be a model for other diseases and foundations. And the prospects for AI/ML are so bright and the field is moving so quickly that I expect the investment Target ALS is making with this initiative will pay enormous dividends now and in the future.”
And the potential doesn’t stop there. Partnerships with AI/ML companies could bring additional expertise and acceleration, providing an opportunity to download the images and apply their algorithms to study TDP-43 pathology.
“This is the largest and first initiative to generate stained and digitized images in ALS,” noted Dr. Harris. “Pairing them with machine learning and sequencing data has the potential to transform how we study the disease.”
Opening the Doors to Discovery
For researchers worldwide, the impact is immediate. Instead of requesting “cases with lots of TDP-43” and relying on someone else’s judgment, they can explore the images directly in the Data Engine, define “lots” for themselves, and request precisely the samples they need.
This democratization of access, rich, standardized datasets that combine imaging, sequencing, and clinical metadata, has long been a bottleneck in ALS research. Now, thanks to this collaboration, the field has a scalable blueprint for the future. Once validated, these AI tools can be deployed at virtually no cost, even as web-based platforms. That’s especially promising for regions of the world with few neuropathologists, making advanced ALS pathology research accessible far beyond well-resourced labs.
The Bigger Picture
Dr. Finkbeiner also noted that once neuropathology is digitized and computable, researchers can integrate it with genomic, transcriptomic, proteomic, and clinical data. This could reveal how genetic risk translates into tissue changes and, ultimately, symptoms and disease progression; insights that will sharpen therapeutic targets and improve the predictive power of preclinical models.
TDP-43 isn’t just another protein. It’s one of the central threads connecting sporadic and familial ALS, as well as frontotemporal dementia and other neurodegenerative diseases. By digitizing and opening these resources, Target ALS is helping the research community visualize pathology in a new way, one that can be integrated with genetics, clinical data, and even animal models to speed up therapeutic discovery.
In the words of Dr. Bowser, “Making these images freely available is about empowering the research community. This initiative is going to be very fruitful, not just for our labs, but for anyone looking to understand and treat ALS.”
Your support helps Target ALS continue building the tools and resources that accelerate discovery. Join us in fueling breakthroughs, donate today.


